



This part of the tutorial introduces you to proteins. HIV protease is a protein that is involved in the production of mature viral proteins in HIV-infected cells. It catalyzes the hydrolysis of peptide bonds in the precursor protein to give a number of functional proteins, one of which is the HIV protease. Proteins with catalytic properties are called enzymes. Because the structure and function of HIV protease are very different from the structure and function of mammalian proteases, this protein is a good target for anti-HIV drugs. Many of the current HIV drugs, such as saquinavir, act by inhibiting the activity of HIV protease. However, because the HIV RNA-based genome is very susceptible to random mutations, the protease is constantly evolving to counter the drugs. The design of new drugs to inhibit such evolving protein remains an important goal of the pharmaceutical industry. Drug design is greatly facilitated by visualization and analysis of active sites of target proteins.
You will download the structure of HIV protease with the bound inhibitor saquinavir from an online database. The main database for protein structures is the RCSB Protein Data Bank, also known as the PDB, and its address is www.rcsb.org. Each structure in the Protein Data Bank has a unique 4-character alphanumeric identifier known as the PDB ID. You can retrieve all related structures using search by keyword, or access a particular structure by its PDB ID.
Search the PDB by keyword for 'HIV Protease'. The search will find nearly two hundred structures, most of which differ by the chemical nature of the bound inhibitor. Narrow your search by specifying the inhibitor you we are interested in. The structure we want is 1FB7.
The entry looks like this:
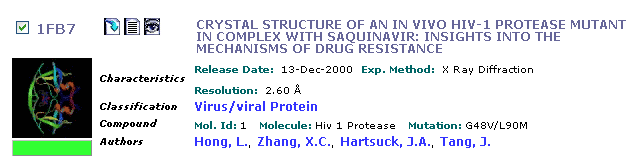
Click on the title and inspect the description for this structure. Notice that a link to the PubMed is provided in the Primary Citation field; these links are often valuable when you need to learn more about a particular protein. Next, download the coordinate file. To download the coordinate file, click on Download Files link on the menu bar and save the PDB text file into your directory (Right-click, then Save File As...).
Open the structure file 1FB7 with SYBYL:
In the 'Edit' menu, select 'Clear All'
In the 'File' menu, select 'Open'
Select file 1FB7.pdb in the your directory, and hit OK to open the file.
Examine the molecule closely. Rotate the molecule until you find the N-terminal residue of this polypeptide. Now rotate the molecule so that you can identify the bound inhibitor; you can recognize the inhibitor easily because it contains quinoline and decahydroquinoline moieties that is not found in proteins. The inhibitor is structurally similar to the normal substrate of the HIV protease, and it binds at a location known as the active site.
Assignments (HIV Protease):As you just saw, macromolecular structures can be quite complex, and sometimes a simplified representation of the molecule is more useful than a detailed atomic representation. One simplified way to present protein structures is by their secondary structures. Main secondary structural elements in proteins are α-helices, β-sheets, and loops. To render the secondary structure of this protein follow these steps:
In the 'View' menu, under 'Biopolymer Display', select 'Ribbon/Tube '
Select everything by clicking on the button marked All and then hit OK
Another window pops up; answer OK to color the protein by the secondary structure
In the 'View' select 'Undisplay Atoms...'
Click on the button marked Substructures, select ROC39, an HIV protease inhibitor and click OK
Click Invert to select everything but the inhibitor, and hit OK
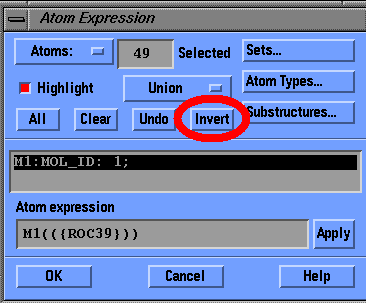
Assignments (HIV Protease Continued):
The structure you are seeing here is only one of two polypeptide chains that make up the active HIV protease. You will now continue to study the active site of dimeric HIV protease in more detail.

